Current projects in the lab
Host factors regulating persistence.
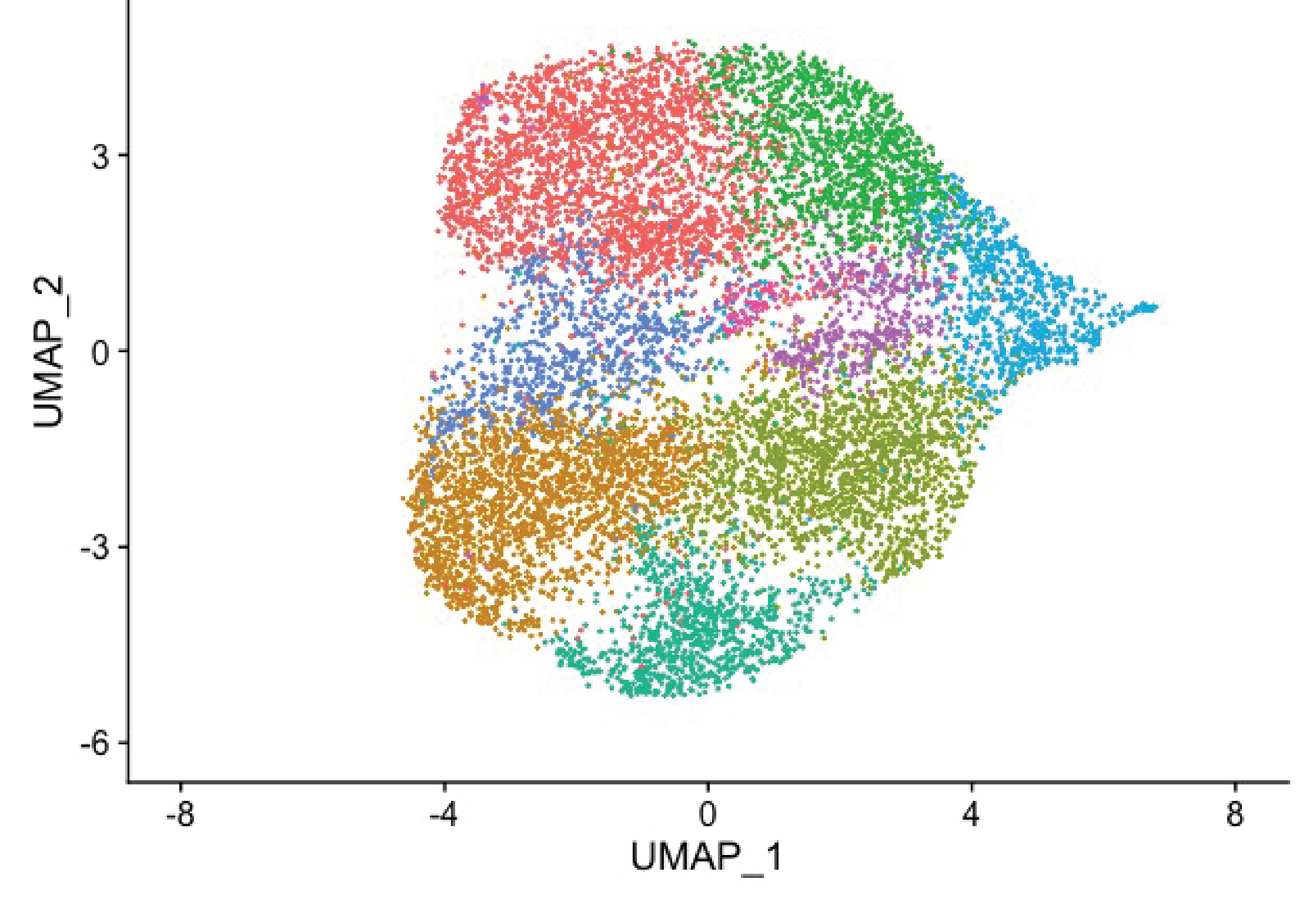
Until recently, it has been challenging to study the HPV lifecycle following infection. We and others have optimized infection of patient-derived primary cells to investigate how host-virus interactions contribute to initial genome amplification during the establishment phase and long-term viral persistence. Whether a virus persists could (1) be purely stochastic, (2) depend on some pre-existing cellular state (e.g., baseline activation of innate immunity), (3) rely on differential viral gene expression, or (4) (likely) a combination of these. To investigate this heterogeneity in outcomes, we used single-cell RNA Seq to interrogate the transcriptional profile of 5000 infected primary human cervical-derived keratinocytes with HPV18. We identified hundreds of human genes differentially regulated in HPV18 infected cells. To my knowledge, we are the only lab in the world using single-cell RNA-Seq to study how the virus manipulates individual cells. The paper describing these data is currently in preparation. We are currently following up on one of the identified targets. Since an inhibitor for this pathway is currently in clinical trials for non-HPV tumors, this project has immediate translational potential. This research is the focus of a funded Arizona Biomedical Research Centre (ABRC) grant, a new NIAID R01 grant, and a pending NIAID R21 proposal . This project also illustrates our lab philosophy to translate large scale genomics data into actionable biochemistry and cell biology research.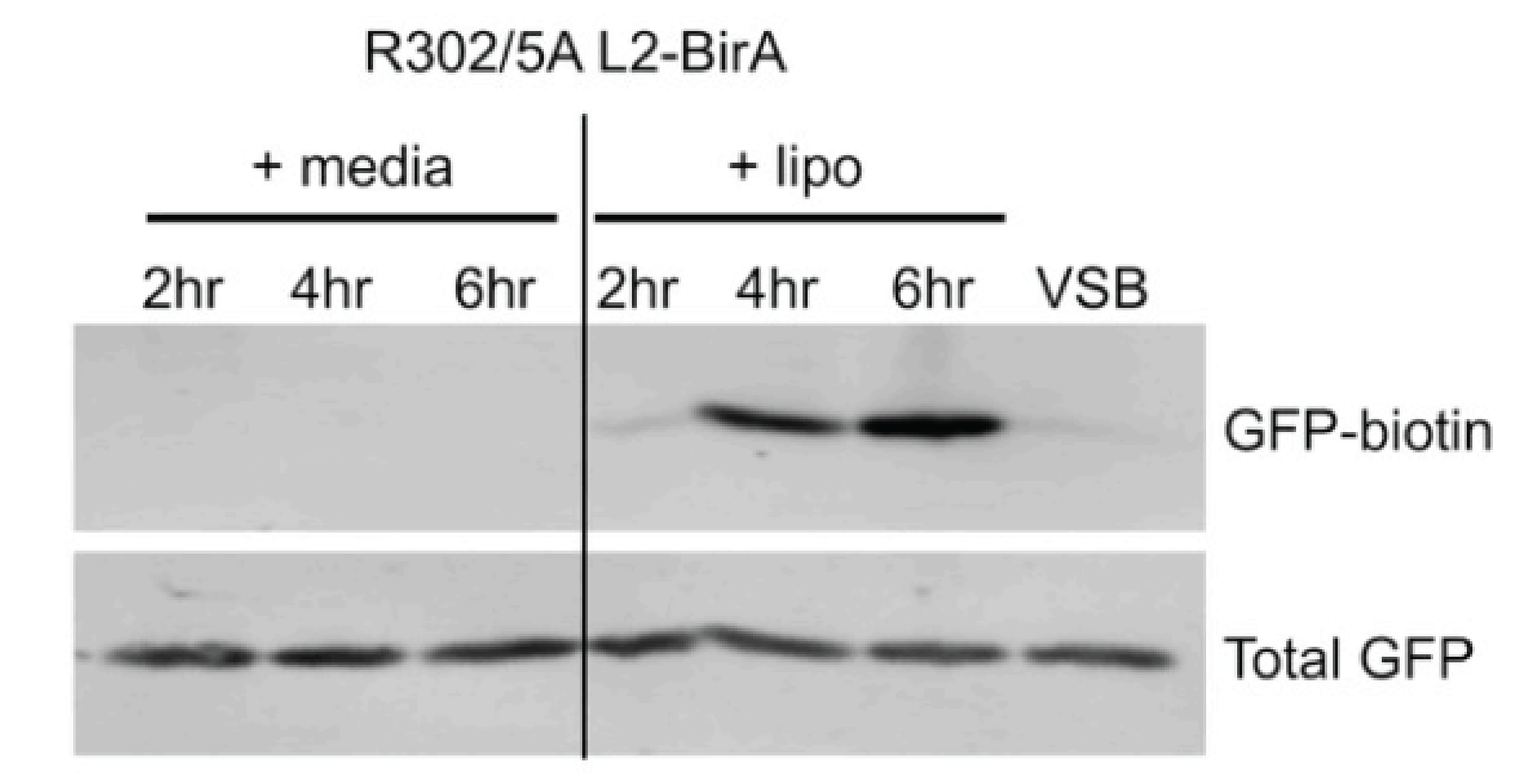
Role of the innate immune system during persistent infection.
Cellular innate immune pathways exist to restrict viral infections. Innate immune responses are initiated after recognizing molecular products associated with microbial infections, known as pathogen-associated molecular patterns (PAMPs). More recent data have illustrated that in addition to PAMPs, DNA generated from cellular damage (host damage-associated molecular patterns (DAMPs)) activate the innate immune response. In collaboration with the lab of Dr. Campos, my lab studies how HPV manipulates the cGAS/STING arm of the innate immune response. Cytosolic DNA activates the cGAS/STING pathway. This initiates a signaling cascade that results in an antiviral response, detrimental to the establishment and persistence of HPV infections. The virus must have developed mechanisms to counteract cellular cGAS/STING responses. Dr. Campos and I have published a paper ( (Uhlorn et al., 2020) on this pathway (Co-corresponding authors) and are preparing another manuscript to present this data. This work is supported by a new American Cancer Society grant to my lab. Dr. Campos and I have received funding to support this project in our labs. We also have a pending R01 grant studying these interactions. We are also interested in how papillomaviruses have evolved to evade detection by TLR9 and APOBEC3. In recently published papers (Warren et al., 2015 and King et al., 2022), we demonstrate how interaction with these innate immune sensors is impacting the evolutionary history of papillomaviruses. Indeed, we believe that the host-virus interactions are critical in driving the evolution of papillomaviruses. Interestingly, this evolutionary pressure appears to be primarily acting at the level of the viral DNA without altering viral protein composition.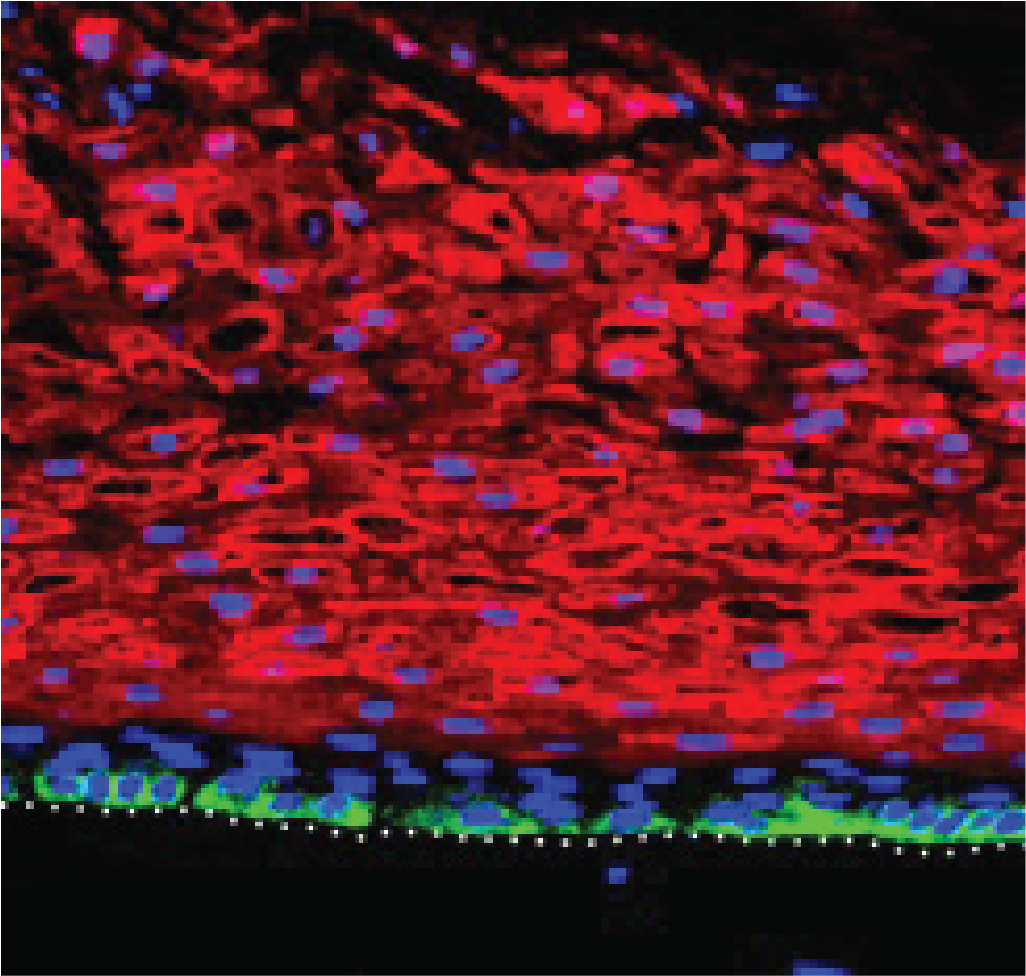
Viral manipulation of cellular differentiation.
The ability to persist in actively dividing cells is essential to the viral lifecycle and oncogenicity. While a delay of cellular differentiation is critical, completion of the viral lifecycle depends on the terminal differentiation of the cell. Indeed, a differentiation-dependent mechanism ensures that 'late' structural genes are not expressed until viral replication has occurred. During keratinocyte differentiation, Spatio-temporal changes in gene expression determine epidermis structure and function. HPVs have evolved to alter this balance while intricately linking their lifecycle to this physiological process. These processes are poorly understood on the host and the viral side. Our overall goal is to determine the critical host-virus interactions that regulate the late stages of a differentiation-dependent HPV lifecycle.We use a physiologically relevant system, HPV16 positive tonsillar epithelium equivalents grown in 3D organotypic raft culture , to characterize the cellular genes regulating Spatio-temporal control of late viral gene expression. Specifically, we are characterizing the transcriptome of individual cells derived from these epidermises using a single-cell RNASeq approach to characterize the host genes whose expression is highly correlated with late viral gene expression. In parallel, we are performing a genome-wide CRISPR/Cas9 knockout screen in these 3D organotypic raft cultures. Since our screening approach targets differentiated cells, this genome-wide genetic dropout screen will determine which cellular genes are crucial for late viral gene transcription. This NIDCR R03 funded research will generate multiple testable hypotheses addressing the dramatic increase in HPV-positive head and neck cancers in future R01 level proposals.
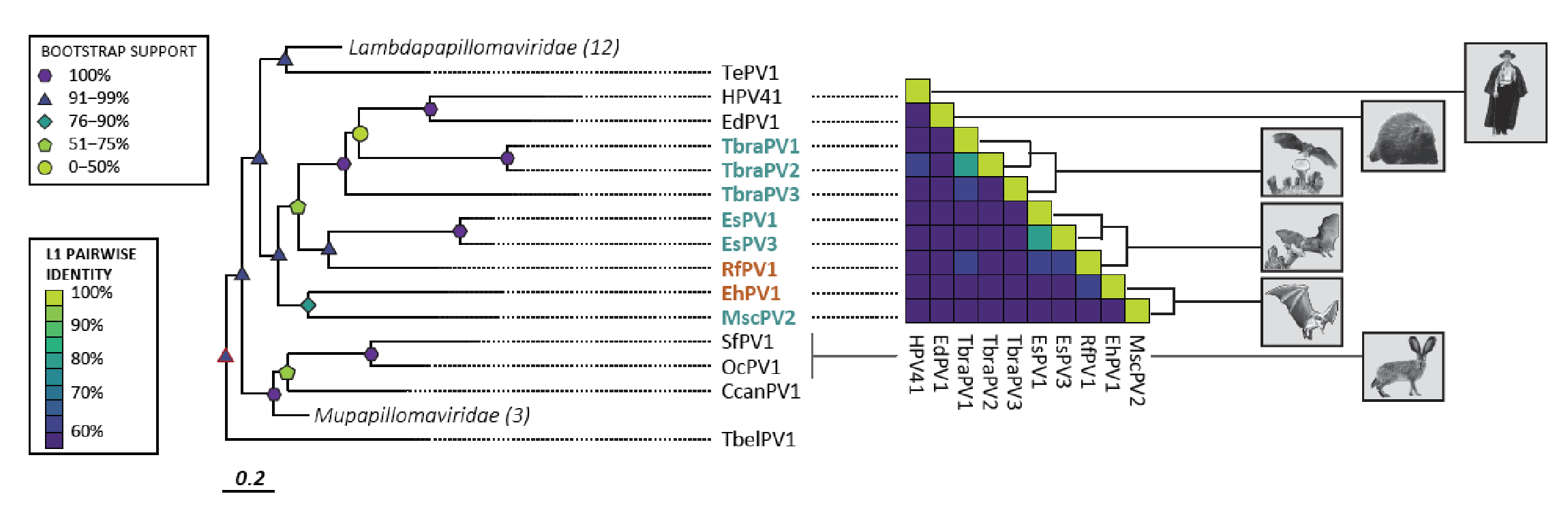
Virus Discovery and Evolution.
As the COVID19 pandemic illustrates, it is crucial to understand how viruses evolve. This requires that we know what viruses circulate in humans and animals. The discovery of novel viruses has been a substantial effort by my laboratory in collaboration with the lab of Dr. Varsani. We sample a wide array of animal and human tissues and use Next-Generation sequencing to identify the viruses in these samples. New viruses are further characterized and used to study the evolutionary history of the ‘virome’ (the total collection of viruses). We published 15 papers on this topic. This research is supported by seed funding from the BIO5 institute and a USDA NIFA award to my lab.
